Reading Zarr files with R package stars
- What is Zarr?
- Aggregating global precipitation time series to countries
- simple aggregation
- CMIP6 data
- Software versions
Summary: Zarr files are the new cloud native NetCDF, and are here to stay. This blog post will explore how they can be read in R using the GDAL “classic” raster API and the (relatively) new GDAL multidimensional array API, and how we can read sub-arrays or strided (subsampled, lower resolution) arrays. CMIP6 data is being released as Zarr data on the Google Cloud.
What is Zarr?
Zarr is a data format; it does not come in a single file as NetCDF
or HDF5 does but as a directory with chunks of data in compressed
files and metadata in JSON files. Zarr was developed in the Python numpy
and xarray communities, and was quickly taken up by the Pangeo
community. A Python-independent specification of Zarr (in progress, V3)
is found here.
GDAL has a Zarr driver, and can read single (spatial, raster) slices without time reference through its classic raster API, and full time-referenced arrays through its newer multidimensional array API. In this blog post we show how these can be used through R and package stars for raster and vector data cubes. We will start with an attempt to reproduce what Ryan Abernathey did with Python, xarray and geopandas, published here.
Aggregating global precipitation time series to countries
Ryan’s blog post aggregates a global, 1-degree “square” gridded daily precipitation dataset (NASA GPCP) to country values, where the aggregation should be weighted according to the amount of overlap of grid cells (as “square” polygons) and country polygons.
We can read the rainfall data in R using
library(stars)
## Loading required package: abind
## Loading required package: sf
## Linking to GEOS 3.10.2, GDAL 3.4.3, PROJ 8.2.0; sf_use_s2() is TRUE
dsn = 'ZARR:"/vsicurl/https://ncsa.osn.xsede.org/Pangeo/pangeo-forge/gpcp-feedstock/gpcp.zarr"'
bounds = c(longitude = "lon_bounds", latitude = "lat_bounds")
r = read_mdim(dsn, bounds = bounds)
r
## stars object with 3 dimensions and 1 attribute
## attribute(s), summary of first 1e+05 cells:
## Min. 1st Qu. Median Mean 3rd Qu. Max.
## precip [mm/d] 0 0 0 2.2841 1.618025 103.5965
## dimension(s):
## from to offset delta refsys point values x/y
## longitude 1 360 0 1 NA NA NULL [x]
## latitude 1 180 -90 1 NA NA NULL [y]
## time 1 9226 NA NA POSIXct NA 1996-10-01,...,2021-12-31
st_bbox(r)
## xmin ymin xmax ymax
## 0 -90 360 90
r[r < 0 | r > 100] = NA
where
- the dsn is constructed such that GDAL knows this is a Zarr file
(
ZARR:) and that it is a web resource (/vsicurl/). - we specify the bounds because the
latitudeandlongitudevariables do not have aboundsattribute pointing to the cell boundaries, and their values do not point to grid cell centers (as recommended but not required by CF 1.10), but to boundaries - the Zarr has no CRS, so that R wouldn’t know these are ellipsoidal coordinates; without action it would then assume they are Cartesian coordinates (as geopandas does); we will correct this later on
- values below 0 and above 100 are set to
NA; this is according to thevalid_rangeattribute for theprecipvariable, but not automatically parsed timedoes not have an offset/delta pair, indicating it is not regular; there seem to be a few duplicate time steps, we can identify them by
time(r) |> diff() |> table()
##
## 0 86400
## 3 9222
ti = time(r)
ti[which(diff(ti) == 0)]
## [1] "2017-08-01 UTC" "2017-11-01 UTC" "2018-04-01 UTC"
50m natural Earth country boundaries
We can load the 50m NE Country boundaries from the rnaturalearth
package:
library(rnaturalearth)
ne = ne_countries(scale = 50, returnclass = "sf")
st_crs(r) = st_crs(ne)
ne = st_make_valid(ne)
where
- we set the CRS of the precipitation data cube, equal to that of NE (WGS84)
- we call
st_make_valid()to make the country boundaries valid on the sphere
simple aggregation
aggregate takes the grid cells as points:
(a = aggregate(r, ne, mean, na.rm = TRUE))
## stars object with 2 dimensions and 1 attribute
## attribute(s), summary of first 1e+05 cells:
## Min. 1st Qu. Median Mean 3rd Qu. Max. NA's
## precip [mm/d] 0 3.206655e-05 0.5487042 2.849516 3.355176 97.14838 28631
## dimension(s):
## from to offset delta refsys point
## geometry 1 241 NA NA +proj=longlat +datum=WGS8... FALSE
## time 1 9226 NA NA POSIXct NA
## values
## geometry MULTIPOLYGON (((-69.91182 1...,...,MULTIPOLYGON (((31.42949 -2...
## time 1996-10-01,...,2021-12-31
and returns a vector data cube: an array with 241 country geometries x
9226 time steps (days). We can select the time series for Italy and plot
it, using the time series package xts, and aggregation to monthly mean
values from xts:
library(xts)
## Loading required package: zoo
##
## Attaching package: 'zoo'
## The following objects are masked from 'package:base':
##
## as.Date, as.Date.numeric
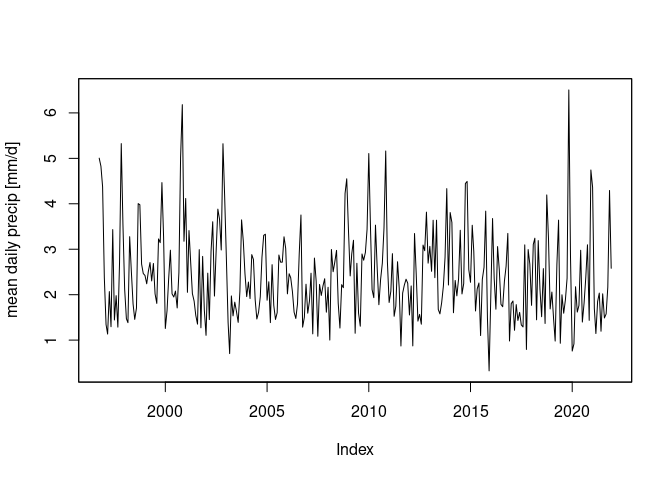
a[, which(ne$admin == "Italy"),] |> # select Italy
adrop() |> # drop geometry dimension
as.xts() -> a.xts # convert to xts (time series class)
a.xts |>
aggregate(as.Date(cut(time(a.xts), "month")), mean, na.rm = TRUE) |> # temporal aggregation
plot(ylab = "mean daily precip [mm/d]")
The “exact” (or “conservative region”) aggregation in Ryan’s blog is
obtained by area-weighted
interpolation;
we have s2 on to get intersection using spherical coordinates; doing
this with Cartesian coordinates will not work for half of the globe
unless we’d recenter the precipitation coordinates from [0,360] to
[-180,180]:
sf_use_s2() # the default
## [1] TRUE
ne = st_make_valid(ne)
it = ne[ne$admin == "Italy",]
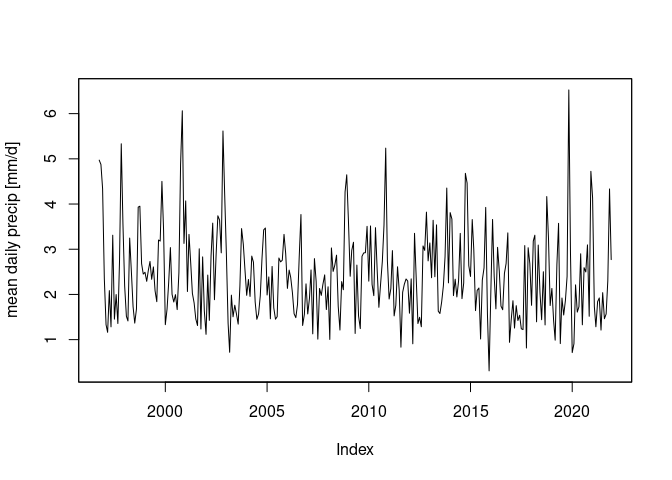
st_interpolate_aw(r, it, extensive = FALSE) |>
st_set_dimensions("time", time(r)) -> a2
## Warning in st_interpolate_aw.sf(st_as_sf(x), to, extensive, ...):
## st_interpolate_aw assumes attributes are constant or uniform over areas of x
a2 |>
adrop() |>
as.xts() |>
aggregate(as.Date(cut(time(a2), "month")), mean, na.rm = TRUE) |> # monthly means
plot(ylab = "mean daily precip [mm/d]")
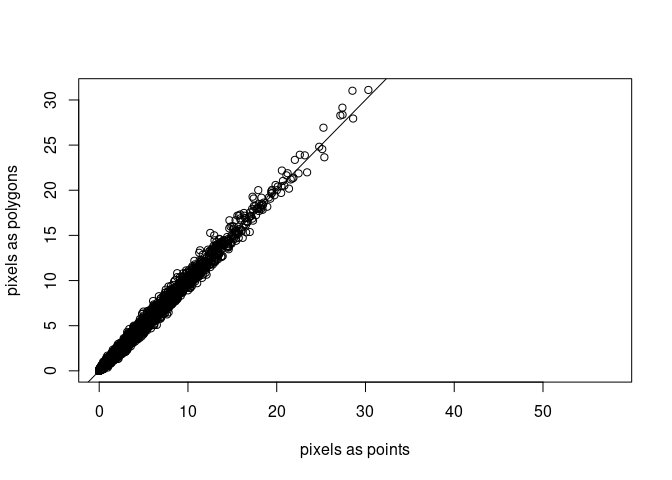
We can compare outcomes from both computations e.g. in a scatter plot:
a2 |> adrop() |> as.xts() -> a2.xts
plot(as.vector(a.xts), as.vector(a2.xts),
xlab = "pixels as points", ylab = "pixels as polygons")
abline(0, 1)
CMIP6 data
CMIP6, the climate model intercomparison project, will share its output data (simulations of climate projections) as NetCDF but also as Zarr files on Google Cloud Storage. A STAC interface (static) to these data is found here.
One of these files, containing pressure at sea level for 6-hour intervals, was created by the following Python script:
import fsspec
import xarray
import zarr
m = fsspec.get_mapper("https://storage.googleapis.com/cmip6/CMIP6/HighResMIP/CMCC/CMCC-CM2-HR4/highresSST-present/r1i1p1f1/6hrPlev/psl/gn/v20170706")
ds = xarray.open_zarr(m)
ds.sel(time=slice("1948-01-01","1955-12-31")).to_zarr("./psl.zarr")
That file is provided here, but we will read the same file directly from the cloud storage using R:
dsn = 'ZARR:"/vsicurl/https://storage.googleapis.com/cmip6/CMIP6/HighResMIP/CMCC/CMCC-CM2-HR4/highresSST-present/r1i1p1f1/6hrPlev/psl/gn/v20170706"/:psl.zarr/'
d = read_mdim(dsn, count = c(NA, NA, 11680))
st_crs(d) = 'OGC:CRS84'
d
## stars object with 3 dimensions and 1 attribute
## attribute(s), summary of first 1e+05 cells:
## Min. 1st Qu. Median Mean 3rd Qu. Max.
## psl [Pa] 95056.82 99547.8 100975.1 100752.8 101700.6 105274.6
## dimension(s):
## from to offset delta refsys point
## lon 1 288 -0.625 1.25 WGS 84 NA
## lat 1 192 NA NA WGS 84 NA
## time 1 11680 1948-01-01 6 hours PCICt_365 NA
## values x/y
## lon NULL [x]
## lat [-90,-89.5288),...,[89.5288,90) [y]
## time NULL
st_bbox(d)
## xmin ymin xmax ymax
## -0.625 -90.000 359.375 90.000
where:
- in addition to the
ZARR:and/vsicurl/prefixes, we see the array name (psl.zarr) specified - the count for time, corresponding to the time slices, is given,
leaving the ones for longitude and latitude to
NA - dimension information can be obtained using the
gdalmdiminfobinary, or from R using
ret = gdal_utils("mdiminfo", dsn, quiet = TRUE)
jsonlite::fromJSON(ret)$dimensions
## name full_name size type direction indexing_variable
## 1 bnds /bnds 2 <NA> <NA> <NA>
## 2 lat /lat 192 HORIZONTAL_Y NORTH /lat
## 3 lon /lon 288 HORIZONTAL_X EAST /lon
## 4 time /time 97820 TEMPORAL <NA> /time
- bounds are being read, and as they are specified as an attribute to the coordinate variables:
jsonlite::fromJSON(ret)$arrays$lon$attributes$bounds
## [1] "lon_bnds"
jsonlite::fromJSON(ret)$arrays$lat$attributes$bounds
## [1] "lat_bnds"
- we can see from the print summary that the
latitudebounds are irregular: the ones touching the poles are half the latitude width as all others. Reading this as a regular GDAL file creates problems. - we also see is the the
refsys(reference system) of the time isPCICt_365, indicating a “365-day” calendar is used: leap days are ignored.
A regular sequence of 6-hour images is provided for the non-leap days.
Using package PCICt we can convert this to POSIXct time (the one we
live in) by
as.POSIXct(r)
## stars object with 3 dimensions and 1 attribute
## attribute(s), summary of first 1e+05 cells:
## Min. 1st Qu. Median Mean 3rd Qu. Max. NA's
## precip [mm/d] 0 0 0 2.283087 1.617958 99.58805 1
## dimension(s):
## from to offset delta refsys point
## longitude 1 360 0 1 +proj=longlat +datum=WGS8... NA
## latitude 1 180 -90 1 +proj=longlat +datum=WGS8... NA
## time 1 9226 NA NA POSIXct NA
## values x/y
## longitude NULL [x]
## latitude NULL [y]
## time 1996-10-01,...,2021-12-31
after which we see that we no longer have a regular time dimension but
semi-regular with gaps (the leap days); POSIXct values are now in the
values field of the time dimension.
We can plot the first 49 time slices e.g. by
plot(r[,,,1:49]) # first 49 time steps
## downsample set to 2

Reading sub-arrays or strided arrays
Using the multidimensional array API we can read lower reslution versions of the imagery by setting
offsetthe offset (start) of reading (pixels, 0, 0, 0 by default)countthe number of steps to read in each dimensionsstepthe step size in each dimension
As an example, let’s try to read one value per year:
(r = read_mdim(dsn, step = c(1, 1, 365 * 4)))
## stars object with 3 dimensions and 1 attribute
## attribute(s), summary of first 1e+05 cells:
## Min. 1st Qu. Median Mean 3rd Qu. Max.
## psl [Pa] 94926.56 99828.71 101017.5 100794.7 101698.9 105274.6
## dimension(s):
## from to offset delta refsys point
## lon 1 288 -0.625 1.25 NA NA
## lat 1 192 NA NA NA NA
## time 1 67 1948-01-01 365 days PCICt_365 NA
## values x/y
## lon NULL [x]
## lat [-90,-89.5288),...,[89.5288,90) [y]
## time NULL
plot(r)
## downsample set to 2

We can for instance read yearly data at a quarter of the spatial resolution by
(r = read_mdim(dsn, step = c(4, 4, 365 * 4)))
## stars object with 3 dimensions and 1 attribute
## attribute(s):
## Min. 1st Qu. Median Mean 3rd Qu. Max.
## psl [Pa] 93488.44 99865.04 101041 100815.3 101760.1 106605
## dimension(s):
## from to offset delta refsys point
## lon 1 72 -2.5 5 NA NA
## lat 1 48 NA NA NA NA
## time 1 67 1948-01-01 365 days PCICt_365 NA
## values x/y
## lon NULL [x]
## lat [-90,-89.5288),...,[86.70157,87.64398) [y]
## time NULL
plot(r)

Using the classic GDAL raster API
We can explore the the zarr file by omitting the array name, e.g. by
dsn = 'ZARR:"/vsicurl/https://storage.googleapis.com/cmip6/CMIP6/HighResMIP/CMCC/CMCC-CM2-HR4/highresSST-present/r1i1p1f1/6hrPlev/psl/gn/v20170706"/'
gdal_utils("info", dsn)
## Driver: Zarr/Zarr
## Files: none associated
## Size is 512, 512
## Subdatasets:
## SUBDATASET_1_NAME=ZARR:"/vsicurl/https://storage.googleapis.com/cmip6/CMIP6/HighResMIP/CMCC/CMCC-CM2-HR4/highresSST-present/r1i1p1f1/6hrPlev/psl/gn/v20170706/":/lat
## SUBDATASET_1_DESC=Array /lat
## SUBDATASET_2_NAME=ZARR:"/vsicurl/https://storage.googleapis.com/cmip6/CMIP6/HighResMIP/CMCC/CMCC-CM2-HR4/highresSST-present/r1i1p1f1/6hrPlev/psl/gn/v20170706/":/lon
## SUBDATASET_2_DESC=Array /lon
## SUBDATASET_3_NAME=ZARR:"/vsicurl/https://storage.googleapis.com/cmip6/CMIP6/HighResMIP/CMCC/CMCC-CM2-HR4/highresSST-present/r1i1p1f1/6hrPlev/psl/gn/v20170706/":/time
## SUBDATASET_3_DESC=Array /time
## SUBDATASET_4_NAME=ZARR:"/vsicurl/https://storage.googleapis.com/cmip6/CMIP6/HighResMIP/CMCC/CMCC-CM2-HR4/highresSST-present/r1i1p1f1/6hrPlev/psl/gn/v20170706/":/lat_bnds
## SUBDATASET_4_DESC=Array /lat_bnds
## SUBDATASET_5_NAME=ZARR:"/vsicurl/https://storage.googleapis.com/cmip6/CMIP6/HighResMIP/CMCC/CMCC-CM2-HR4/highresSST-present/r1i1p1f1/6hrPlev/psl/gn/v20170706/":/lon_bnds
## SUBDATASET_5_DESC=Array /lon_bnds
## SUBDATASET_6_NAME=ZARR:"/vsicurl/https://storage.googleapis.com/cmip6/CMIP6/HighResMIP/CMCC/CMCC-CM2-HR4/highresSST-present/r1i1p1f1/6hrPlev/psl/gn/v20170706/":/psl
## SUBDATASET_6_DESC=Array /psl
## SUBDATASET_7_NAME=ZARR:"/vsicurl/https://storage.googleapis.com/cmip6/CMIP6/HighResMIP/CMCC/CMCC-CM2-HR4/highresSST-present/r1i1p1f1/6hrPlev/psl/gn/v20170706/":/time_bnds
## SUBDATASET_7_DESC=Array /time_bnds
## Corner Coordinates:
## Upper Left ( 0.0, 0.0)
## Lower Left ( 0.0, 512.0)
## Upper Right ( 512.0, 0.0)
## Lower Right ( 512.0, 512.0)
## Center ( 256.0, 256.0)
which lists the subdatasets; if we would ask for info on a subdatasetname we see an error:
dsn = 'ZARR:"/vsicurl/https://storage.googleapis.com/cmip6/CMIP6/HighResMIP/CMCC/CMCC-CM2-HR4/highresSST-present/r1i1p1f1/6hrPlev/psl/gn/v20170706/":/psl'
gdal_utils("info", dsn)
## Warning in CPL_gdalinfo(if (missing(source)) character(0) else source,
## options, : GDAL Error 1: Indices of extra dimensions must be specified
indicating that we need to add an index of the additional dimension (time). If we add e.g. index 10, we get
dsn = 'ZARR:"/vsicurl/https://storage.googleapis.com/cmip6/CMIP6/HighResMIP/CMCC/CMCC-CM2-HR4/highresSST-present/r1i1p1f1/6hrPlev/psl/gn/v20170706/":/psl:10'
gdal_utils("info", dsn)
## Driver: Zarr/Zarr
## Files: none associated
## Size is 288, 192
## Origin = (-0.625000000000000,-90.471204188481678)
## Pixel Size = (1.250000000000000,0.942408376963351)
## Metadata:
## cell_measures=area: areacella
## cell_methods=area: time: mean
## comment=Sea Level Pressure
## long_name=Sea Level Pressure
## standard_name=air_pressure_at_sea_level
## Corner Coordinates:
## Upper Left ( -0.6250000, -90.4712042)
## Lower Left ( -0.6250000, 90.4712042)
## Upper Right ( 359.375, -90.471)
## Lower Right ( 359.375, 90.471)
## Center ( 179.3750000, 0.0000000)
## Band 1 Block=288x192 Type=Float32, ColorInterp=Undefined
## NoData Value=1.00000002004087734e+20
## Unit Type: Pa
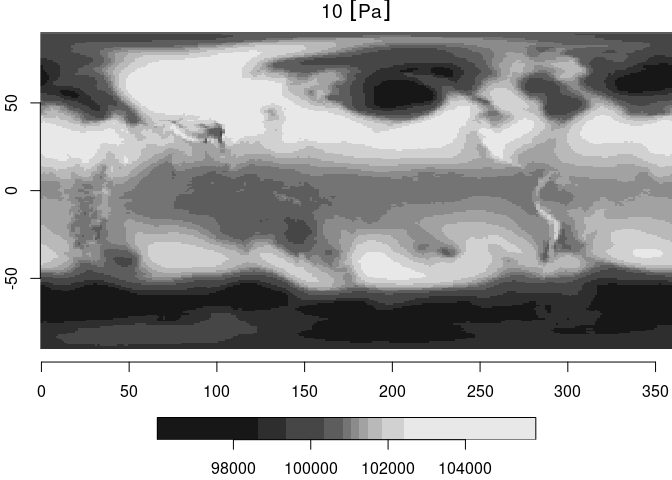
which describes a 288 x 192 pixel raster layer. It also reveals that
GDAL thinks that this raster file has latitude dimensions exceeding
[-90,90]. We can read such a slice with read_stars and plot the
result:
(r0 = read_stars(dsn))
## stars object with 2 dimensions and 1 attribute
## attribute(s):
## Min. 1st Qu. Median Mean 3rd Qu. Max.
## 10 [Pa] 96032.01 99854.94 101043.6 100784.3 101660.2 105820.4
## dimension(s):
## from to offset delta refsys point values x/y
## x 1 288 -0.625 1.25 NA NA NULL [x]
## y 1 192 -90.4712 0.942408 NA NA NULL [y]
plot(r0, axes = TRUE)
Software versions
To use the features in this blog post for reading sub-arrays or lower resolution arrays, you need stars >= 0.5-7 and sf >= 1.0-9, which you can get by installing from source by
remotes::install_github("r-spatial/sf") # requires rtools42 on windows
remotes::install_github("r-spatial/stars")
You need both, since stars uses sf’s binding to GDAL - stars is a
pure R package, all linking to GDAL happens through sf.
Alternatively, and in particular if you use Windows or MACOS you could install the github versions as binary package from r-universe:
install.packages("sf", repos = "https://r-spatial.r-universe.dev")
install.packages("stars", repos = "https://r-spatial.r-universe.dev")
update users have reported
failure to reproduce
the steps in this blog post. Note that this blog post was run on a
version of sf linked to GDAL 3.4.3 from the
ubuntugis-unstable ppa. If you
have sf linked to an older version of GDAL, or if the GDAL version was
not linked against BLOSC, then this (or parts of it) may not work. Note
that Zarr, and Zarr support in GDAL are all very recent developments,
and it takes time for modern library support to propagate through
e.g. CRAN build toolchains – we are working on it!
