Spatial indexes coming to sf
Spatial indexes give you fast results on spatial queries,
such as finding whether pairs of geometries intersect
or touch, or finding their intersection. They reduce
the time to get results from quadratic in the number of
geometries to linear in the number of geometries. A recent
commit
brings spatial indexes to sf for the binary logical predicates
(intersects, touches, crosses, within, contains,
contains_properly, overlaps, covers, covered_by,
relate_pattern, equals, disjoint), as well as intersection,
which returns intersecting geometries.
The spatial join function st_join uses a logical predicate to
join features, and is also also affected by this speedup.
Antecedents
There have been attempts to use spatial planar indices, including
enhancement issue sfr:76.
In rgeos, GEOS STRtrees were used in
rgeos/src/rgeos_poly2nb.c, which is mirrored in a modern Rcpp setting
sf/src/geos.cpp, around lines 276 and 551.
The STRtree is constructed by building envelopes (bounding boxes) of input entities,
which are then queried for intersection with envelopes of another set of entities
(in rgeos, R functions gUnarySTRtreeQuery and gBinarySTRtreeQuery). The use case
was to find neighbours of all the about 90,000 US Census entities in Los Angeles, via
spdep::poly2nb(), which received an argument to enter the candidate neighbours found
by Unary querying the STRtree of entities by the same entities.
Benchmark
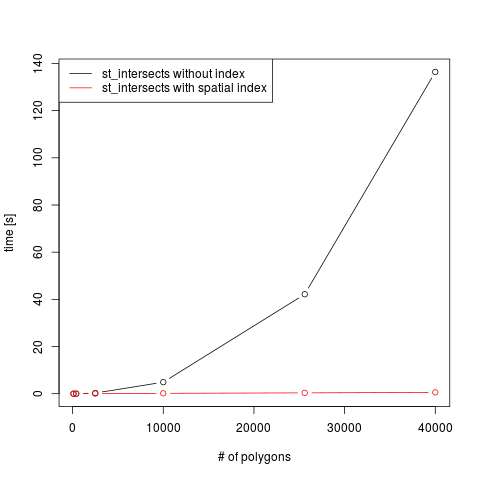
A simple benchmark shows the obvious: st_intersects without spatial
index behaves quadratic in the number of geometries (black line),
and is much faster for the case where a spatial index is created,
stronger so for larger number of polygons:
The polygon datasets used are simple checker boards with square polygons (showing a nice Moiré pattern):
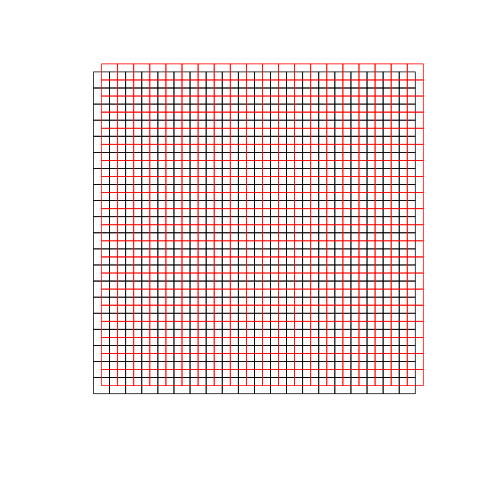
The black small square polygons are essentially matched to the red ones; the number of polygons along the x axis is the number of a single geometry set (black).
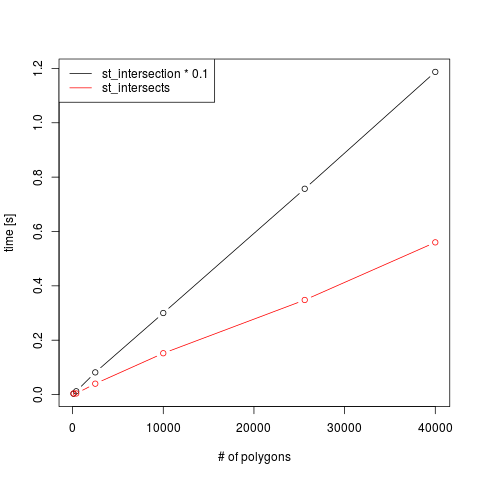
To show that the behaviour of intersects and intersection
is indeed linear in the number of polygons, we show runtimes for
both, as a function of the number of polygons (where intersection
was divided by 10 for scaling purposes):
Implementation
Spatial indexes are available in the
GEOS library used by sf, through the
functions
starting with STRtree. The algorithm
implements a Sort-Tile-Recursive R-tree, according to the JTS
documentation
described in P. Rigaux, Michel Scholl and Agnes Voisard. Spatial
Databases With Application To GIS. Morgan Kaufmann, San Francisco,
2002.
The sf implementation
(some commits to follow this one) excludes some binary operations.
st_distance, and st_relate without pattern, are excluded as
these need to go through all combinations, returning a dense
matrix. st_equals_exact is excluded as approximate matches are
sought, without constraining the degree of approximation.
On which argument is an index built?
The R-tree is built on the first argument (x), and used to
match all geometries over the second argument (y) of binary
functions. This could give runtime differences, but for instance
for the dataset that triggered this development in
sfr:394, we see hardly
any difference:
library(sf)
# Linking to GEOS 3.5.1, GDAL 2.1.3, proj.4 4.9.2, lwgeom 2.3.2 r15302
load("test_intersection.Rdata")
nrow(test)
# [1] 16398
nrow(hsg2)
# [1] 6869
system.time(int1 <- st_intersection(test, hsg2))
# user system elapsed
# 105.712 0.040 105.758
system.time(int2 <- st_intersection(hsg2, test))
# user system elapsed
# 107.756 0.060 107.822
# Warning messages:
# 1: attribute variables are assumed to be spatially constant throughout all geometries
# 2: attribute variables are assumed to be spatially constant throughout all geometries
The resulting feature sets int1 and int2 are identical, only
the order of the features (records) and of the attribute columns
(variables) differs. Runtime without index is 35 minutes, 20 times
as long.
Is the spatial index always built?
In the current implemenation (version >= 0.5-1) the index is always
built, except for st_distance, and st_relate without pattern.
This means it is also built e.g. for rook or queen neighborhood
selections (sfr:234),
which use st_relate with a specified pattern.
What about prepared geometries?
Prepared geometries in GEOS are essentially indexes over single geometries and not over sets of geometries; they speed things up in particular when single geometries are very complex, and only for a single geometry to single geometry comparison. The spatial indexes are indexes over collections of geometries; they make a cheap preselection based on bounding boxes before the expensive pairwise comparison takes place.
Script used
The followinig script was used to create the benchmark plots. It no longer works; in the version where it worked, prepared = FALSE would take a branch where no trees were built, this is no longer the case.
library(sf)
sizes = c(10, 20, 50, 100, 160, 200)
res = matrix(NA, length(sizes), 4)
for (i in seq_along(sizes)) {
g1 = st_make_grid(st_polygon(list(rbind(c(0,0),c(0,1),c(1,1),c(0,1),c(0,0)))), n = sizes[i]) * sizes[i]
g2 = g1 + c(.5,.5)
res[i, 1] = system.time(i1 <- st_intersects(g1, g2))[1]
res[i, 2] = system.time(i2 <- st_intersects(g1, g2, prepare = FALSE))[1]
res[i, 3] = system.time(i1 <- st_intersection(g1, g2))[1]
res[i, 4] = identical(i1, i2)
}
plot(sizes^2, res[,2], type = 'b', ylab = 'time [s]', xlab = '# of polygons')
lines(sizes^2, res[,1], type = 'b', col = 'red')
legend("topleft", lty = c(1,1), col = c(1,2), legend = c("st_intersects without index", "st_intersects with spatial index"))
plot(sizes^2, res[,3]/10, type = 'b', ylab = 'time [s]', xlab = '# of polygons')
lines(sizes^2, res[,1], type = 'b', col = 'red')
legend("topleft", lty = c(1,1), col = c(1,2), legend = c("st_intersection * 0.1", "st_intersects"))
