mapedit - updates in 0.2.0
mapedit has progressed substantially since the introduction to
mapedit post.
mapedit 0.2.0 offers improvements and incorporates changes based on
much appreciated feedback from the R geospatial community. mapedit is
still in rapid development, but the API is stabilizing. We are targeting
a CRAN release prior to useR 2017, and Tim
Appelhans will demonstrate mapedit in his useR
talk.
In this post, we will highlight some of the recent improvements and
changes to mapedit. These updates can be categorized as
-
better integration with [simple features(https://github.com/edzer/sfr)] and
-
addition of Shiny modules.
Install
We are moving quickly, so please install the development versions of
mapview, leaflet.extras, and mapview as shown below.
devtools::install_github("r-spatial/mapview@develop")
devtools::install_github("bhaskarvk/leaflet.extras")
devtools::install_github("r-spatial/mapedit")
Simple Features
The R geo community is radidly embracing the RConsortium-sponsored sf
package, and mapedit plans to fully adopt and incorporate simple
features like leaflet, mapview, geojsonio, plotly, and
ggplot2. sf can greatly improve geospatial workflows in R. mapedit
now returns simple features by default with editMap() and includes a
new function selectFeatures() for interactive selection of simple
features. Let’s take a quick look at this new functionality.
editMap returns sf
editMap() looks the same, but the output is very different.
library(mapedit)
library(mapview)
library(sf)
crud <- editMap(mapview())
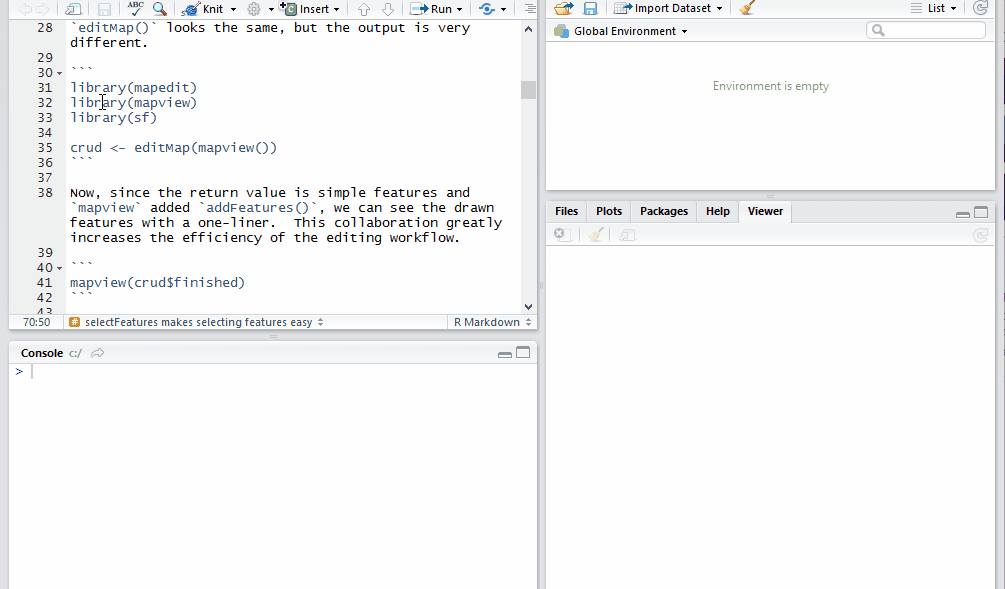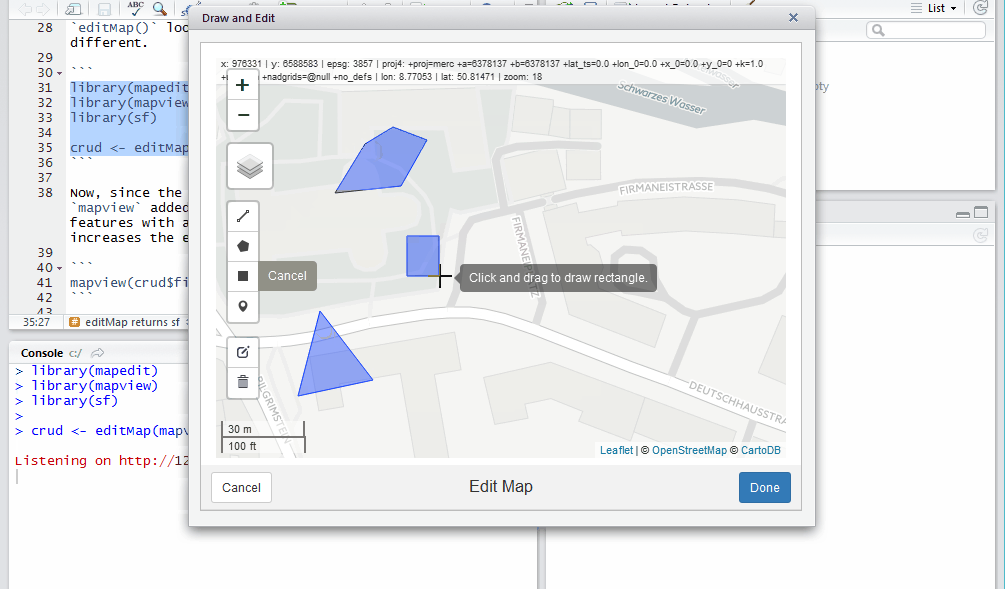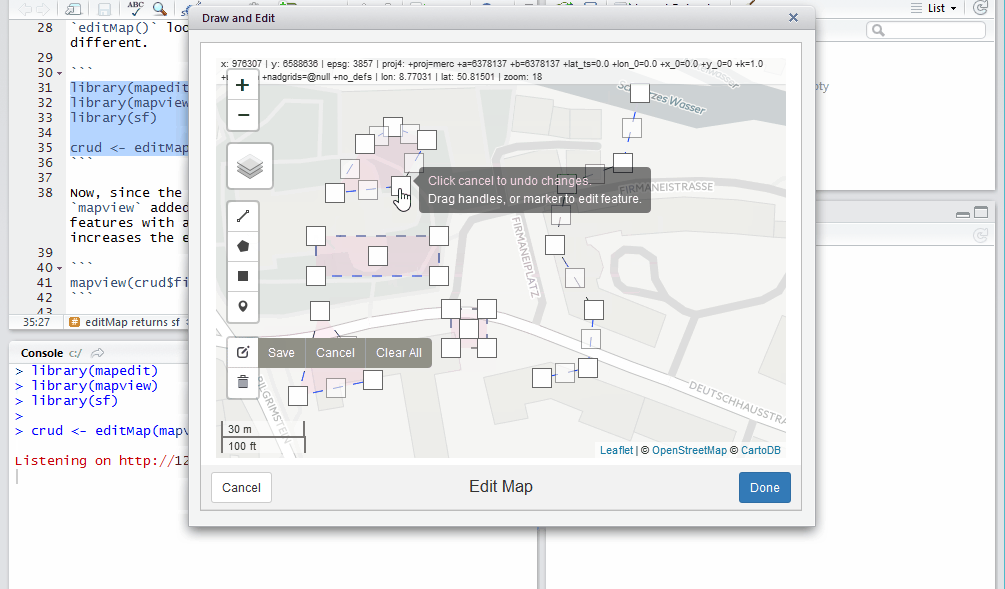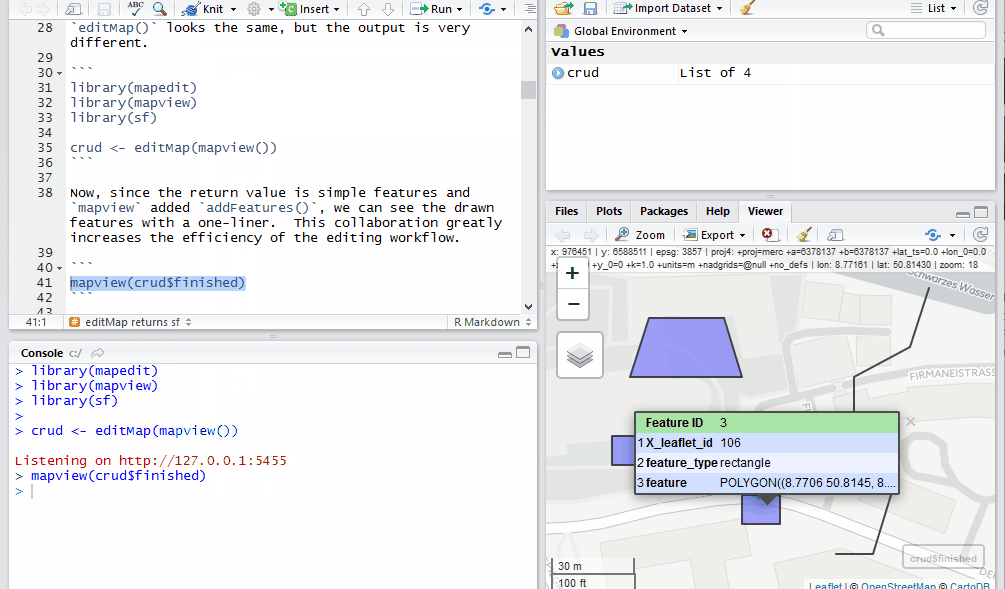
Now, since the return value is simple features and mapview added
addFeatures(), we can see the drawn features with a one-liner. This
collaboration greatly increases the efficiency of the editing workflow.
mapview(crud$finished)
selectFeatures makes selecting features easy
Let’s use the sf example with North Carolina county data to give us
some simple features to select with the new selectFeatures().
library(mapview)
library(mapedit)
library(sf)
nc <- st_read(system.file("shape/nc.shp", package="sf"))
selected <- selectFeatures(nc)
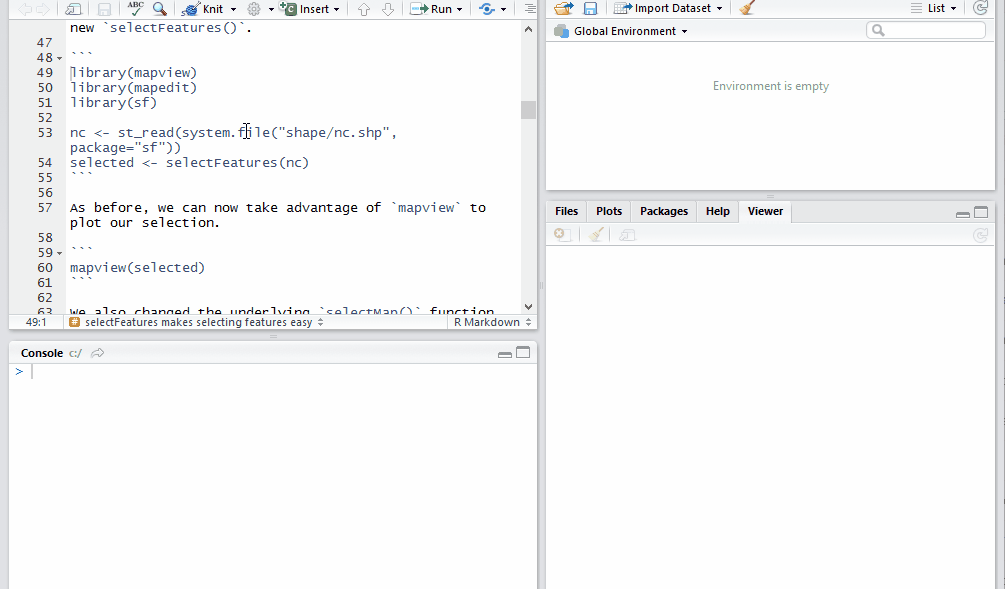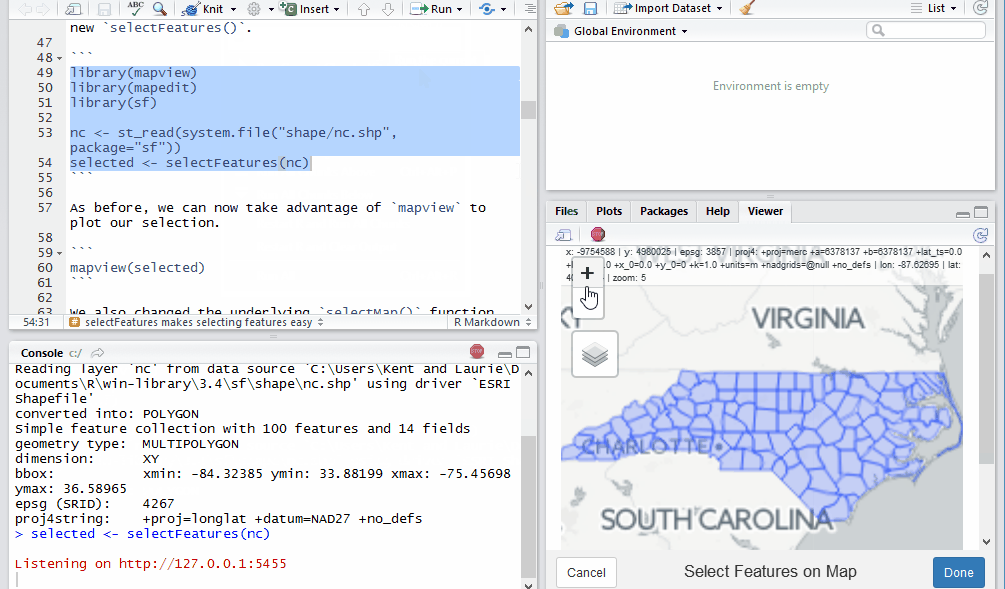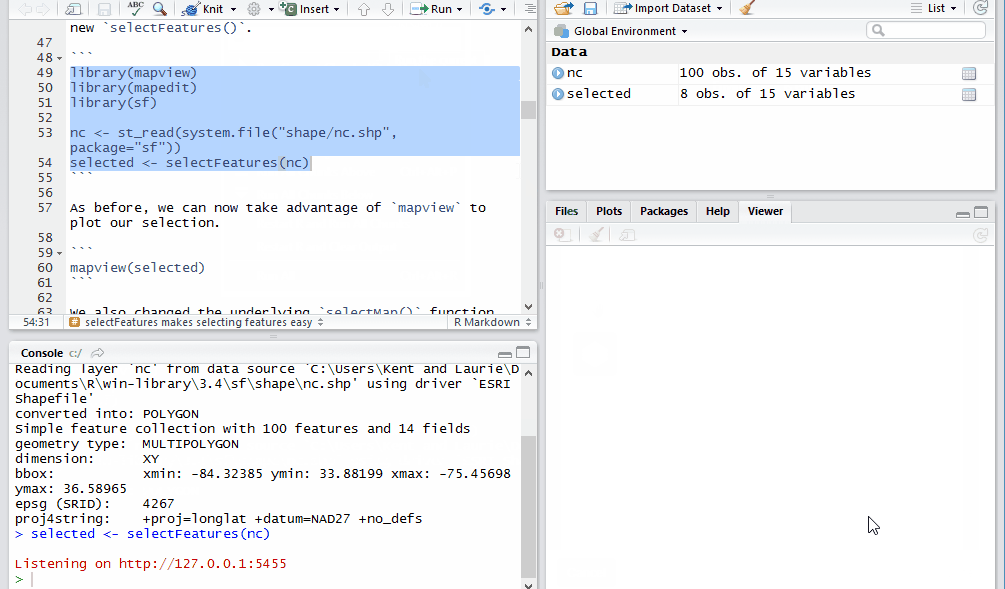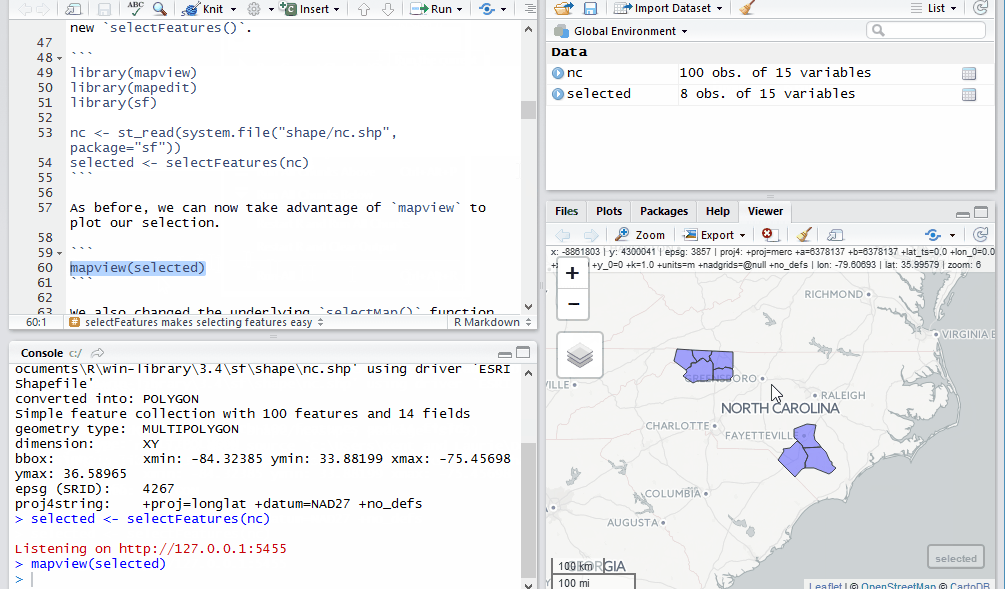
As before, we can now take advantage of mapview to plot our selection.
mapview(selected)
We also changed the underlying selectMap() function to use the RStudio
Viewer by default. This allows us to include selectMap() in a workflow
or pipeline.
library(mapview)
library(mapedit)
library(sf)
nc <- st_read(system.file("shape/nc.shp", package="sf"))
selectFeatures(nc) %>%
st_union() %>%
mapview()
Stay tuned for an editFeatures() equivalent.
Shiny Modules
The original editMap() and selectMap() provided useful
functionality. However, they are limited to standalone application. For
even greater integration in an interactive geospatial workflow, Shiny
modules allow a user
to incorporate edit and select in a broader application context. Let’s
see a couple examples of this concept.
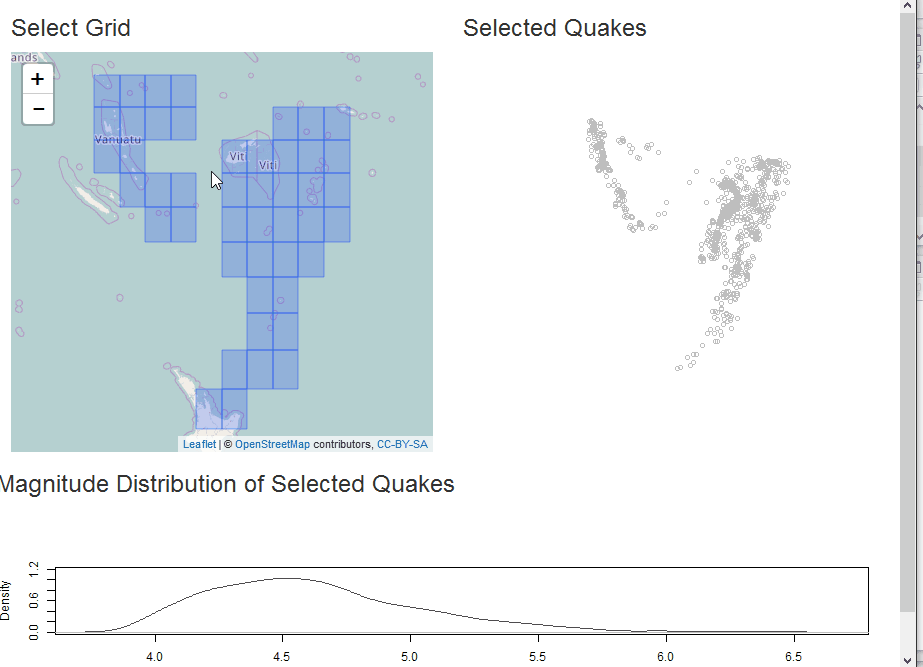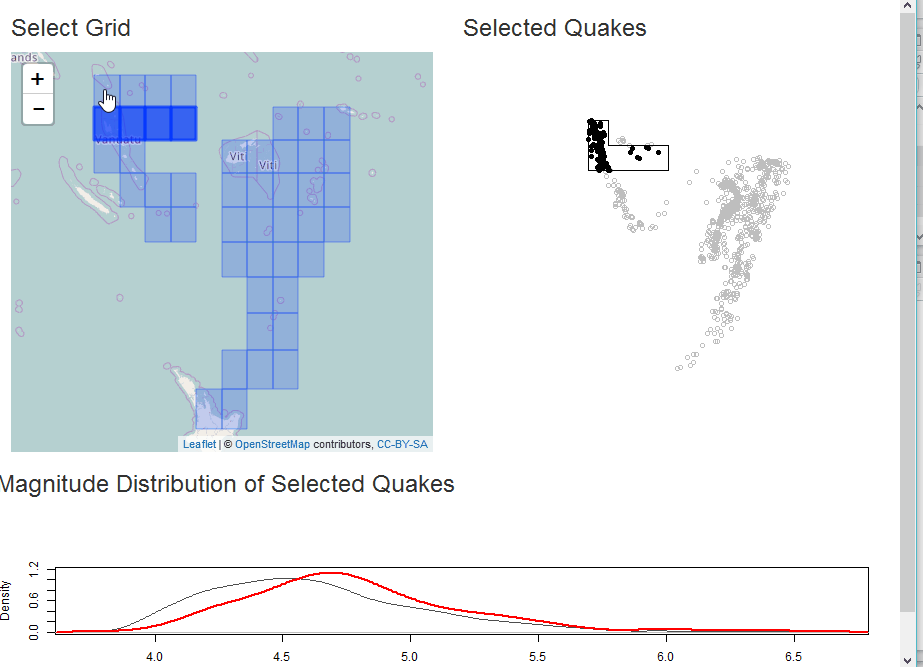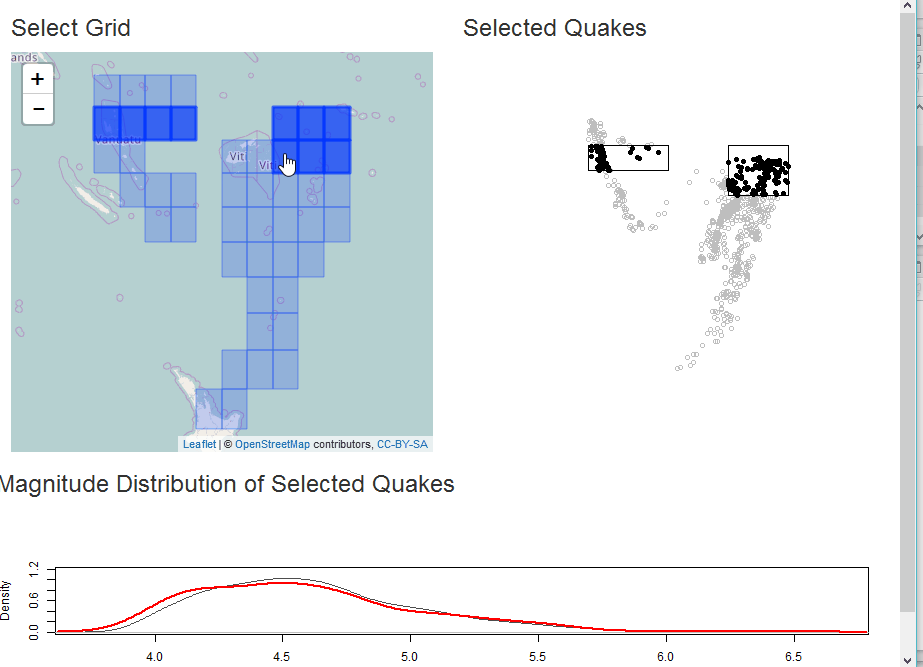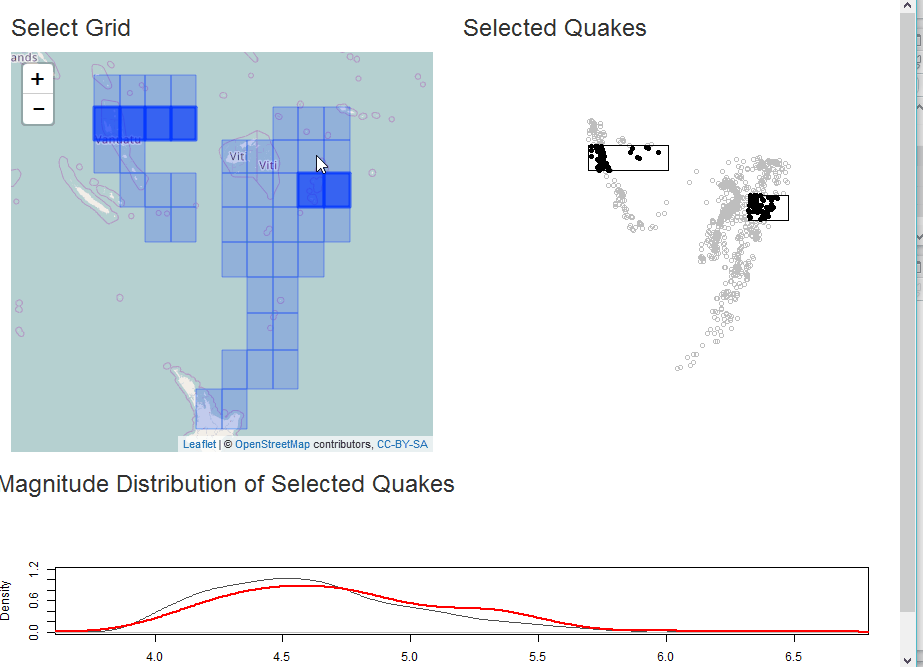
Select as Shiny Module
In this example, we will demonstrate analysis of the quakes data in
R along with some helpful sf. The app will build a grid for
selection of quakes and then plot the selection with a comparative
density plot.
First we will convert the quakes to simple features.
library(sf)
# make the coordinates a numeric matrix
qk_mx <- data.matrix(quakes[,2:1])
# convert the coordinates to a multipoint feature
qk_mp <- st_multipoint(qk_mx)
# convert the multipoint feature to sf
qk_sf <- st_sf(st_cast(st_sfc(qk_mp), "POINT"), quakes, crs=4326)
Now let’s use the very helpful sf::st_make_grid() function, and then
filter the grid to only those that contain quakes points.
# make a grid
grd <- st_set_crs(st_make_grid(qk_sf), 4326)
# only keep grid polygons that contain at least one quake point
grd <- grd[which(sapply(st_contains(st_sf(grd), qk_sf),length)>0)]
With our grid, we can build a Shiny app for some interactive analysis of quake magnitude.
library(mapview)
library(mapedit)
library(shiny)
ui <- fluidPage(
fluidRow(
column(
6,
h3("Select Grid"),
# our new select module ui
selectModUI("selectmap")
),
column(
6,
h3("Selected Quakes"),
plotOutput("selectplot")
)
),
fluidRow(
h3("Magnitude Distribution of Selected Quakes"),
plotOutput("quakestat", height=200)
)
)
server <- function(input, output, session) {
# our new select module
g_sel <- callModule(
selectMod,
"selectmap",
leaflet() %>%
addTiles() %>%
addFeatures(st_sf(grd), layerId = ~seq_len(length(grd)))
)
rv <- reactiveValues(intersect=NULL, selectgrid=NULL)
observe({
# the select module returns a reactive
# so let's use it to find the intersection
# of selected grid with quakes points
gs <- g_sel()
rv$selectgrid <- st_sf(
grd[as.numeric(gs[which(gs$selected==TRUE),"id"])]
)
if(length(rv$selectgrid) > 0) {
rv$intersect <- st_intersection(rv$selectgrid, qk_sf)
} else {
rv$intersect <- NULL
}
})
output$selectplot <- renderPlot({
plot(qk_mp, col="gray")
if(!is.null(rv$intersect)) {
plot(rv$intersect, pch=19, col="black", add=TRUE)
}
plot(st_union(rv$selectgrid), add=TRUE)
})
output$quakestat <- renderPlot({
plot(
stats::density(qk_sf$mag), col="gray30", ylim=c(0,1.2),
main = NA
)
if(!is.null(rv$intersect) && nrow(rv$intersect) > 0) {
lines(stats::density(rv$intersect$mag), col="red", lwd=2)
}
})
}
shinyApp(ui, server)
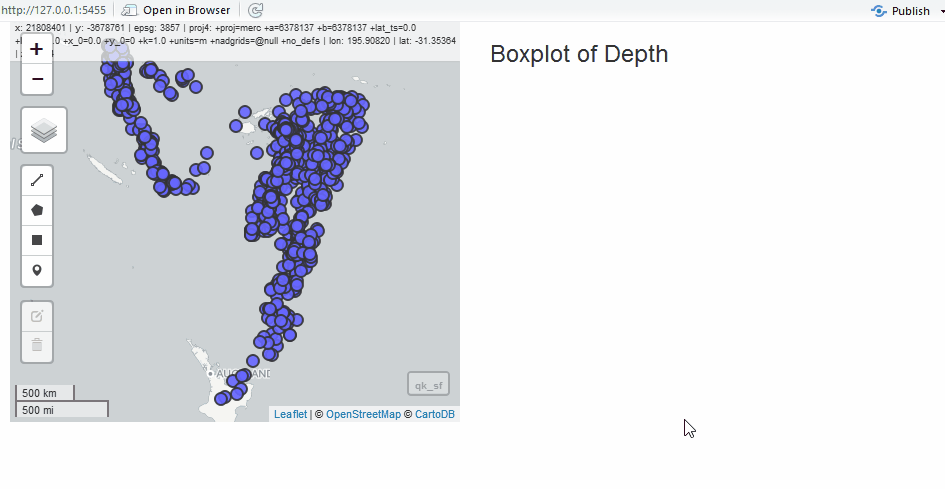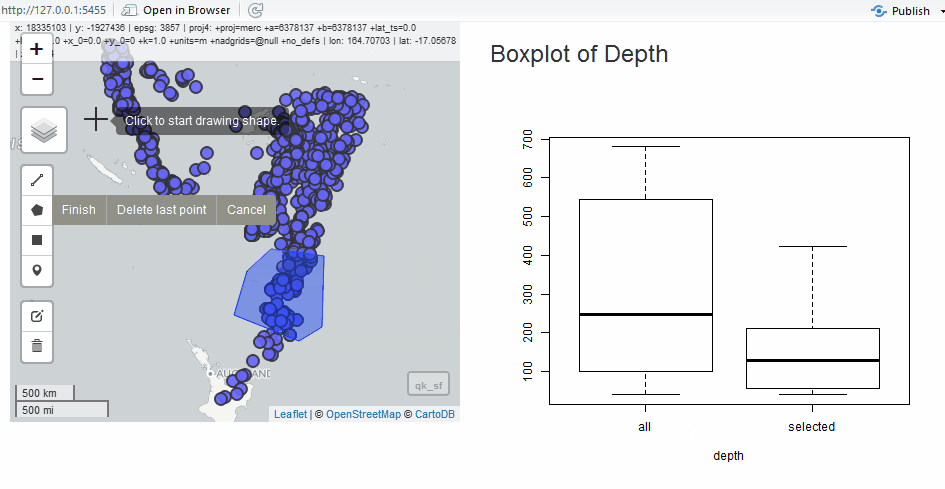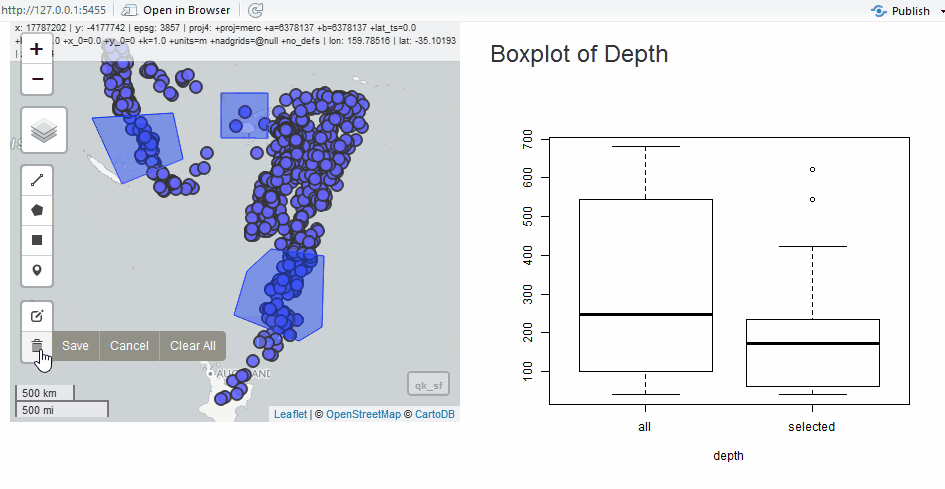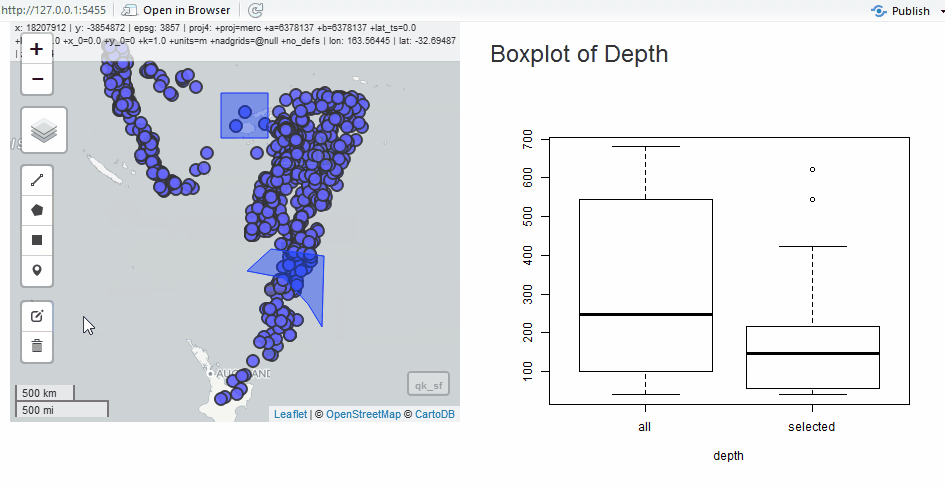
Edit as Shiny Module
Since we have the quake data, we will use it to show the edit module in a simple application. Instead of the grid, let’s draw polygons to select quakes.
# run select demo for the quake data
# we will need the qk_sf
# to test
# plot(qk_sf)
library(mapedit)
library(mapview)
library(shiny)
ui <- fluidPage(
fluidRow(
# edit module ui
column(6, editModUI("editor")),
column(
6,
h3("Boxplot of Depth"),
plotOutput("selectstat")
)
)
)
server <- function(input, output, session) {
# edit module returns sf
edits <- callModule(editMod, "editor", mapview(qk_sf)@map)
output$selectstat <- renderPlot({
req(edits()$finished)
qk_intersect <- st_intersection(edits()$finished, qk_sf)
req(nrow(qk_intersect) > 0)
boxplot(
list(
all = as.numeric(qk_sf$depth),
selected = as.numeric(qk_intersect$depth)
),
xlab = "depth"
)
})
}
shinyApp(ui, server)
Next Steps
The progress made thus far depended entirely on feedback received. Please help us by providing feedback, ideas, and use cases. As mentioned earlier, we aim for an initial CRAN release before useR 2017 on July 4, 2017. We do not anticipate any breaking API changes before release. Rather, we plan to spend time on documentation, examples, and tests.
RConsortium
mapedit and many of its dependency packages are funded by the
RConsortium. Thanks so much to all
those who have contributed to this fantastic organization. Also, thanks
to all those open source contributors in the R community.
